Client For :
Helmholtz Munich, Germany
Service :
Summer Research Intern
(DAAD WISE Scholar)
Overview
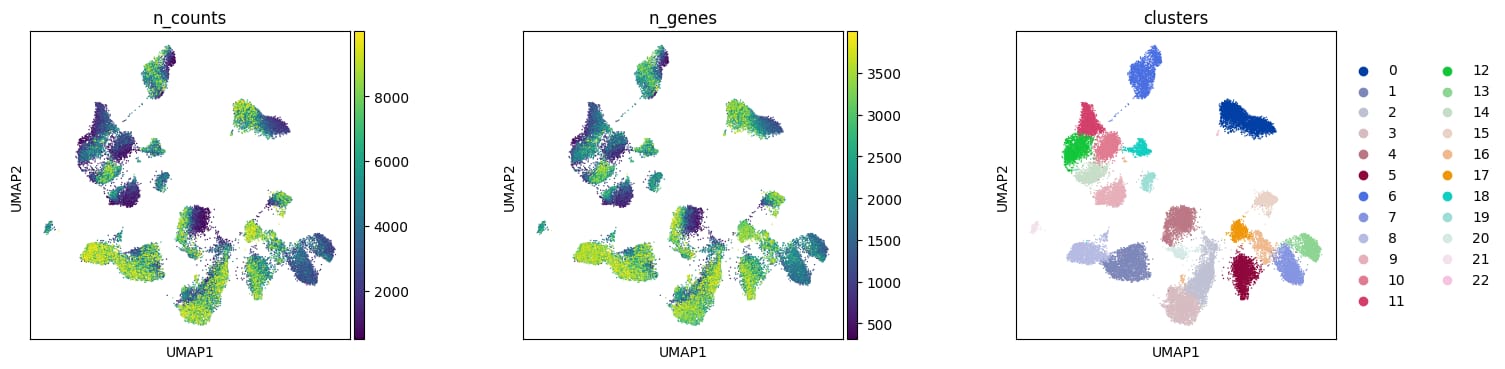
Filtration curves in topological data analysis (TDA) applied to single-cell RNA sequencing (scRNA-seq) data refer to a process of studying the shape and features of high-dimensional gene expression data across multiple scales. This approach uses algebraic topology tools, such as persistent homology, to capture and quantify the multi scale topological features (like connected components, loops, and voids) that persist throughout a sequence of nested spaces generated by varying a filtration parameter.
Challenges
Data Sparsity and Noise: scRNA-seq data is inherently noisy and sparse due to drop-out events and technical variability, complicating the construction of reliable topological representations.
Prototyping and Testing :
Initial prototypes use Mapper algorithm to build topological representations by constructing networks from pairwise correlation distances among highly variable genes.
Stability tests include varying gene sets, filtration parameters, and coverage to assess robustness of the topological features.
Results/Conclusion :
Documentation and user-friendly software packages (e.g., Mapper implementations, scTDA) are provided to facilitate adoption by biomedical researchers.
The research focused on utilizing Filtration Curves for the analysis of cell graphs through spatial data. Our team worked on implementing Graph Descriptor Functions (Assortative and Average path length).